Research project "Kill Asthma"
With a worldwide prevalence of around three percent, asthma is one of the most common chronic diseases. There is no non-communicable disease among children that is more common. Affected people suffer from a narrowing of the bronchi, which can lead to severe shortness of breath and - if treatment is inadequate or inadequate - to death.
It is therefore important to recognize asthma early on in order to give patients - especially children - the opportunity to intervene quickly in the event of an attack and to contain the symptoms. The diagnosis is typically made by assessing the severity of various symptoms and the volume of the lungs; a specific test does not yet exist.
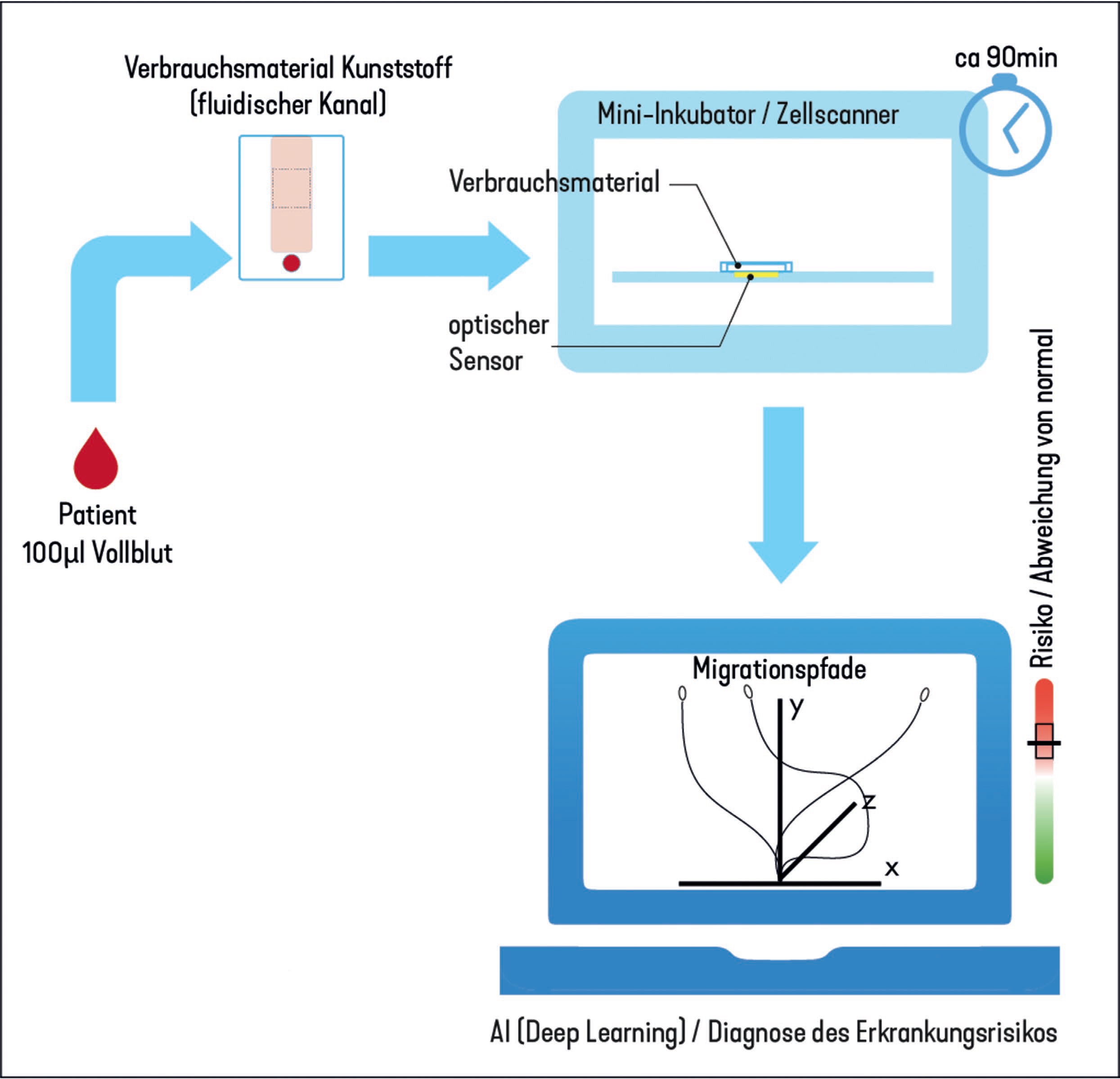
This is where the KillAsthma project comes in, which is funded by the state of Schleswig-Holstein. In collaboration with two high-tech companies from Schleswig-Holstein, the cell processing group is developing a rapid test that only requires a drop of blood - and the immune cells it contains - to diagnose asthma. In 2014, a US group found evidence that asthma is also reflected in changes in the movement patterns of certain immune cells (neutrophil granulocytes).
Within the current project, a holographic microscope has already been designed and built, which enables automatic tracking (tracking) of thousands of cells in three dimensions. It can be miniaturized and produced very cheaply thanks to a lensless design. The movement patterns determined in this way are transferred to an artificial neural network (AI-supported analysis). Hence the acronym of the project: AI-based analysis of immune cells for asthma diagnosis.
After extensive training with data sets marked as "inconspicuous" or "pathological", which are extracted from the blood of real patients, such an AI should be able to detect or rule out asthma in a previously unknown data set with a high degree of certainty. In general, artificial neural networks can learn from the parallel analysis of many training data patterns that are hidden from human perception due to their high abstraction. We hope not only to be able to differentiate clearly between healthy and conspicuous patient data, but also to be able to characterize the individual expression of the highly diverse clinical picture.